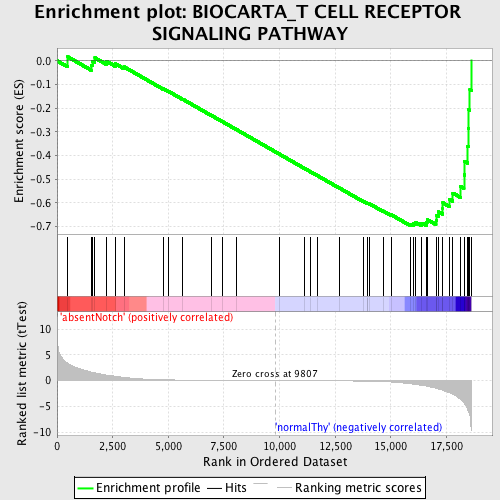
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_absentNotch_versus_normalThy.phenotype_absentNotch_versus_normalThy.cls #absentNotch_versus_normalThy.phenotype_absentNotch_versus_normalThy.cls #absentNotch_versus_normalThy_repos |
| Phenotype | phenotype_absentNotch_versus_normalThy.cls#absentNotch_versus_normalThy_repos |
| Upregulated in class | normalThy |
| GeneSet | BIOCARTA_T CELL RECEPTOR SIGNALING PATHWAY |
| Enrichment Score (ES) | -0.6982937 |
| Normalized Enrichment Score (NES) | -1.587745 |
| Nominal p-value | 0.0039525693 |
| FDR q-value | 0.12524526 |
| FWER p-Value | 0.85 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | GRB2 | 6650398 | 481 | 3.303 | 0.0169 | No | ||
| 2 | PTPRC | 130402 5290148 | 1532 | 1.647 | -0.0183 | No | ||
| 3 | PPP3CB | 6020156 | 1587 | 1.586 | -0.0007 | No | ||
| 4 | MAP2K4 | 5130133 | 1700 | 1.499 | 0.0127 | No | ||
| 5 | RAC1 | 4810687 | 2211 | 1.042 | -0.0013 | No | ||
| 6 | RAF1 | 1770600 | 2612 | 0.832 | -0.0120 | No | ||
| 7 | MAPK8 | 2640195 | 3011 | 0.616 | -0.0255 | No | ||
| 8 | HRAS | 1980551 | 4771 | 0.157 | -0.1182 | No | ||
| 9 | PPP3CA | 4760332 6760092 | 5002 | 0.136 | -0.1288 | No | ||
| 10 | PLCG1 | 6020369 | 5644 | 0.094 | -0.1621 | No | ||
| 11 | ARHGAP6 | 2060121 | 6942 | 0.049 | -0.2313 | No | ||
| 12 | PIK3R1 | 4730671 | 7432 | 0.039 | -0.2571 | No | ||
| 13 | MAP2K1 | 840739 | 8058 | 0.027 | -0.2904 | No | ||
| 14 | ARHGAP5 | 2510619 3360035 | 9991 | -0.003 | -0.3944 | No | ||
| 15 | SOS1 | 7050338 | 11112 | -0.021 | -0.4545 | No | ||
| 16 | JUN | 840170 | 11408 | -0.026 | -0.4700 | No | ||
| 17 | NCF2 | 540129 2370441 2650133 | 11707 | -0.032 | -0.4856 | No | ||
| 18 | PIK3CA | 6220129 | 12692 | -0.056 | -0.5379 | No | ||
| 19 | CAMK2B | 2760041 | 13788 | -0.104 | -0.5955 | No | ||
| 20 | SHC1 | 2900731 3170504 6520537 | 13933 | -0.115 | -0.6018 | No | ||
| 21 | PRKCA | 6400551 | 13968 | -0.118 | -0.6021 | No | ||
| 22 | FOS | 1850315 | 14050 | -0.124 | -0.6048 | No | ||
| 23 | ARHGAP1 | 2810010 5270064 | 14656 | -0.191 | -0.6349 | No | ||
| 24 | NFATC1 | 510400 2320348 4050600 6180161 6290136 6620086 | 15011 | -0.268 | -0.6505 | No | ||
| 25 | FYN | 2100468 4760520 4850687 | 15899 | -0.618 | -0.6903 | Yes | ||
| 26 | ARFGAP1 | 4780364 5390358 | 16026 | -0.687 | -0.6882 | Yes | ||
| 27 | ZAP70 | 1410494 2260504 | 16100 | -0.731 | -0.6826 | Yes | ||
| 28 | PPP3CC | 2450139 | 16390 | -0.935 | -0.6861 | Yes | ||
| 29 | RALBP1 | 4780632 | 16595 | -1.057 | -0.6834 | Yes | ||
| 30 | MAPK3 | 580161 4780035 | 16632 | -1.091 | -0.6712 | Yes | ||
| 31 | ARFGAP3 | 3390717 | 17031 | -1.485 | -0.6734 | Yes | ||
| 32 | PTPN7 | 3450110 | 17065 | -1.530 | -0.6554 | Yes | ||
| 33 | LAT | 3170025 | 17151 | -1.641 | -0.6387 | Yes | ||
| 34 | NFKBIA | 1570152 | 17332 | -1.860 | -0.6243 | Yes | ||
| 35 | MAP3K1 | 5360347 | 17339 | -1.868 | -0.6005 | Yes | ||
| 36 | RELA | 3830075 | 17641 | -2.379 | -0.5859 | Yes | ||
| 37 | LCK | 3360142 | 17786 | -2.667 | -0.5591 | Yes | ||
| 38 | VAV1 | 6020487 | 18137 | -3.653 | -0.5306 | Yes | ||
| 39 | CD4 | 1090010 | 18290 | -4.362 | -0.4823 | Yes | ||
| 40 | CD3D | 2810739 | 18306 | -4.485 | -0.4250 | Yes | ||
| 41 | CD3E | 3800056 | 18448 | -5.536 | -0.3610 | Yes | ||
| 42 | CD247 | 3800725 5720136 | 18488 | -5.985 | -0.2856 | Yes | ||
| 43 | ARHGAP4 | 1940022 | 18504 | -6.111 | -0.2072 | Yes | ||
| 44 | CD3G | 2680288 | 18554 | -6.817 | -0.1216 | Yes | ||
| 45 | TRB@ | 3940292 | 18616 | -9.644 | -0.0000 | Yes |

